The Ubiquitin-Proteasome System
The UPS is key to cellular protein homeostasis, stabilisation, and degradation
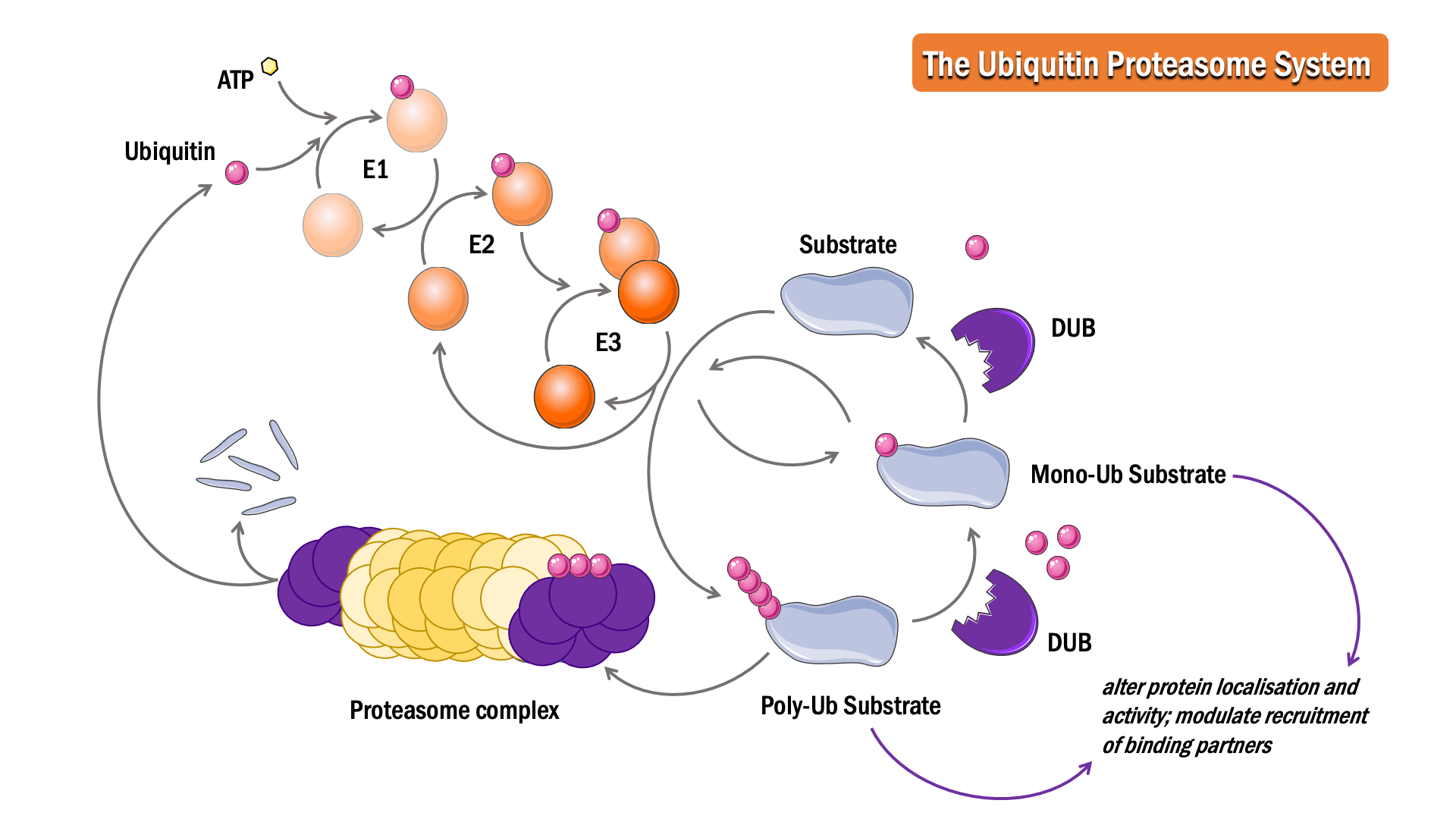
In this process, ubiquitin, a 76 amino acid protein, is attached to a substrate protein, resulting in either mono- or poly-ubiquitination.
The position of attachment and structures of the ubiquitin chains determine whether a protein modulates a specific signalling cascade or is degraded in a proteasomal or lysosomal dependent manner.
The sequential action of three classes of enzymes are required for ubiquitination: activating enzymes (E1s), conjugating enzymes (E2s) and ligases (E3s). In an ATP-dependent first step, E1 binds to and activates ubiquitin, forming a thioester linkage, to initiate degradation. The ubiquitin is then transferred to the sulphydryl group of the active-site cysteine on the E2 enzyme, forming an E2-ubiquitin intermediate. E3 binds both the intermediate and the substrate protein, facilitating an isopeptide bond formation between the ubiquitin and the substrate protein to complete the pathway.
The Deubiquitinase (DUB) enzymes are proteases responsible for deubiquitination. Cleavage of ubiquitin from proteins can prevent protein degradation, alter protein localisation and activity, and modulate the recruitment of binding partners.
DUBs in novel therapeutic applications
The DUB enzymes regulate ubiquitination in the UPS, presenting an exciting new class of drug targets and opportunity for novel modalities to address areas of high unmet medical need.
Dysregulation of deubiquitinase (DUB) activity is implicated in many human diseases, including cancer and neurodegenerative diseases, highlighting their importance and promise for the development of novel therapeutics.
DUBs can be targeted or harnessed for therapeutic effect:
DUB Inhibitors
When DUB activity is the driver of disease pathology, DUB inhibitors can reduce this activity by inhibiting target protein ubiquitination, leading to therapeutic benefits.

DUB-Targeting Proteolysis Targeting Chimeras (PROTACs)
When DUB activity is the driver of disease, but inhibition of the enzyme may be insufficient to achieve a therapeutic effect, DUB-targeting PROTACs can induce the degradation of the DUB. Degradation through the recruitment of E3 ligases to ubiquitinate the DUB target may be able to provide a more sustained reduction in signalling than enzymatic inhibition.

DUB-Targeting Chimeras (DUBTACs)
There are many conditions, including monogenic diseases, where the loss of protein function, such as due to the presence of a genetic mutation, drives disease pathology. DUBTACs could have a therapeutic effect through the recruitment of DUBs to deubiquitinate a target protein, and thereby stabilise the protein or modulate its function.

Though a relatively underexploited area to date, the design and discovery of DUB inhibitors, DUB-targeting PROTACs, and DUBTACs is gaining pace and will be key to the development of powerful new medicines.